Manual
Input formats
There are 3 input formats that TMCrys can process. Examples for all can be found on the Submit page. The sequences can also be uploaded as a text file. The formats are:
- Simple FASTA format either single or multiline.
- FASTA format with the topology of the protein. In this case, CCTOP will not be run on the input sequences. The sequence and the topology should be single lined, thus an entry consistst of 3 lines:
- >header
- sequence
- topology
- Space separated format: the id, sequence and topology of the protein, each separated by a single space. The provided topology will be used for the prediction.
Topology should be the same length as the sequence and give the position of every amino acid of the transmembrane protein. The following letters represents the different locations:
- I=>Inside
- O=>Outside
- M=>Membrane
- S=>Signal
- L=>Re-entrant loop
Running time
The running time of TMCrys depends on the server load and the running time of CCTOP and NetSurfp. The latter two softwares' running times increase with the length of the sequence, a longer sequence can take minutes to be processed.
Sequence batches are limited to 10 sequences at a time, depending on the length of its sequences this usually finishes in a few minutes. You can follow the progress on the waiting page by taking a look at the progress bar.
HTML output
The output of TMCrys is diplayed on the result page in HTML format. An example output can be found here.
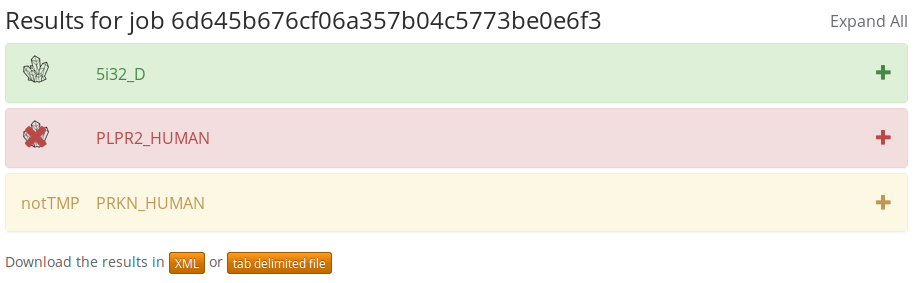
For every transmembrane protein query sequence the following pieces of information are displayed:
- Solubilization/Purification/Crystallization/Whole process: the predicted values of the different crystallization steps as numbers and as a slider diagram. The values are in range [0, 1].
The slider diagram is as follows:
Min value (failure)ThresholdMax value (success)01Predicted valueFigure 2. Example of the slider diagram for the visualization of the prediction.The threshold used for classification is indicated as a yellow stripe between the two sides of the diagram. Different thresholds were set for the different steps. The value of the actual classification is indicated by a blue vertical line. - Sequence: the supplied sequence of the protein.
- Topology: the predicted or supplied topology of the protein.
- Similar proteins in TSTMP
- 3D: transmembrane proteins whose structures are known
- Modelable: transmembrane proteins that could be modeled with existing structures
- Target: transmembrane proteins whose structures are unknown and that do not have structures to modeled with
- Similar proteins in TargetTrack: TargetTrack database incorporated structural genomics experiments with detailed description of methods. Finding similar proteins might help with the process of protein structure determination. Similar proteins are found by running Blast on TargetTrack entries.
- Features: some of the calculated features used for prediction.
Downloadable output formats
The output of TMCrys can be downloaded in multiple formats.
- XML: the schema describing TMCrys output can be downloaded in XSD format.
- Tab separated file: this file consists of 13 columns namely
Namethe name of the query provided by the user in the FASTA headerSolValuepredicted probability for success of solubilizationSolDecisiondecision on the basis of the solubilization value using the defined thresholdPurValuepredicted probability for success of purificationPurDecisiondecision on the basis of the purification value using the defined thresholdCrysValuepredicted probability for success of crystallizationCrysDecisiondecision on the basis of the crystallization value using the defined thresholdWholeValuepredicted probability for success of the whole process from solubilization to crystallizationWholeDecisiondecision on the basis of the whole process value using the defined thresholdRealiabilityreliability of the prediction for the whole processSequencethe sequence of the protein that was provided by the userTopologythe topology of the transmembrane protein calculated by CCTOP or provided by the userTSTMP_3Dsimilar proteins in TSTMP database whose structure has been determined (separated by semicolons)TSTMP_Modelsimilar proteins in TSTMP database that would be able to be modeled from the structure of the query (separated by semicolons)TSTMP_Targetsimilar proteins in TSTMP database that has no known structure and cannot be modeled from existing structures (separated by semicolons)TargetTracksimilar proteins in TargetTrack -experiment IDs with similar protein sequences that might help in the structure determination proces (separated by semicolons).pIisoelectric point of the protein (ProtParam)buriedratioratio of the number of buried and exposed amino acids (NetSurfP)avgRSAaverage solvent accessible surface area averaged across the residues of the protein (NetSurfP)OBlengthLength of sequencegravyGRAVY (ProtParam)lgmwLogarithm of the molecular weight of the proteininindexIntsability index (ProtParam)halfHalf life in mammals (ProtParam)